The arrival of spring often happens quite suddenly: within days, trees can change from bare to bountiful, and the world is filled with flowers!
In a recent publication, Luis Villar and colleagues made a huge step towards understanding the molecular mechanisms that drive floral development in the important crop plant sweet cherry. As well as giving us a deeper understanding of one of the world’s most beautiful natural phenomena, the study also highlights the contrasting importance of both model species research, and that of sometimes focusing beyond our beloved Arabidopsis.
While I’m sure all of us love a world that’s filled with cherry blossoms, Prunus avium, aka sweet cherry, doesn’t flower just to make us happy. And the flower itself, while aesthetically brilliant, is not the valuable product- either for the tree itself, nor, generally, for people. P. avium wants to make fruit- tiny cherry babies wrapped around the seeds of the next generation. And humans (and therefore cherry growers) want ingredients for pie.
Nonetheless, making a flower is an important step in creating that fruit. And unsurprisingly, many of the factors related to floral dud development influence fruit productivity, with changes in floral bud per tree, number of flowers per bud, or simple developmental abnormalities change cherry yield. So understanding floral development, and how, for example, it might be affected by various abiotic changes (we’re looking at you climate change), can be valuable for those of us in the stone-fruit industry, and those of us who happen to like Cherry Ripes*.
(*although, do those even have cherries in them?)
In order to better understand cherry floral development, and ultimately further the flower-field, Villar sampled P. avium floral tissue at different developmental stages, and used RNA sequencing to look at which genes were being expressed at different times. The scientists collected an immense amount of data: 616,862,772 raw sequencing reads from which they could isolate nearly 3000 genes that showed differential expression during floral development. Of course, in order to collect the data, they first had to track that development.
And that process, took nearly nine whole months.
Perhaps unsurprisingly, a lot of what we know about plant flowering comes from the model plant Arabidopsis. More than 90 genes involved in floral regulation have been identified from Arabidopsis, including those that react to endogenous (internal plant) cues, those that respond to environmental factors, and a small set of central regulators that put everything into action.
But Arabidopsis is an annual species, in which the entire floral process occurs in a matter of weeks. For long-living trees, for example cherry, the process can take nearly a year, including a winter period in which the whole tree is dormant!
For cherry, the floral buds that will ultimately give rise to summer fruits first develop nearly a year before those fruits appear. In the first stage (S1), in early spring, the meristematic apex, a section of cells from which new tissues grow, acquires a dome-like structure leading to the formation of flower primordia. In summer, the flower primordia differentiates, and a cup-like structure with 5 sepal primordias at the margin develops (S2). In the S3 stage, the petal, stamen and pistil pimordia develop, and finally, at S4, by the end of summer, the flower has all its whorls present and is ready to go.
Or at least.. ready to go, bar that awkward dormant phase (D), which covers all of autumn and winter. When spring comes back around again, the warming weather signals for the buds to burst forth and fill the world with colour and snow-like petal fall.
Anyway, the process looks a bit like this:
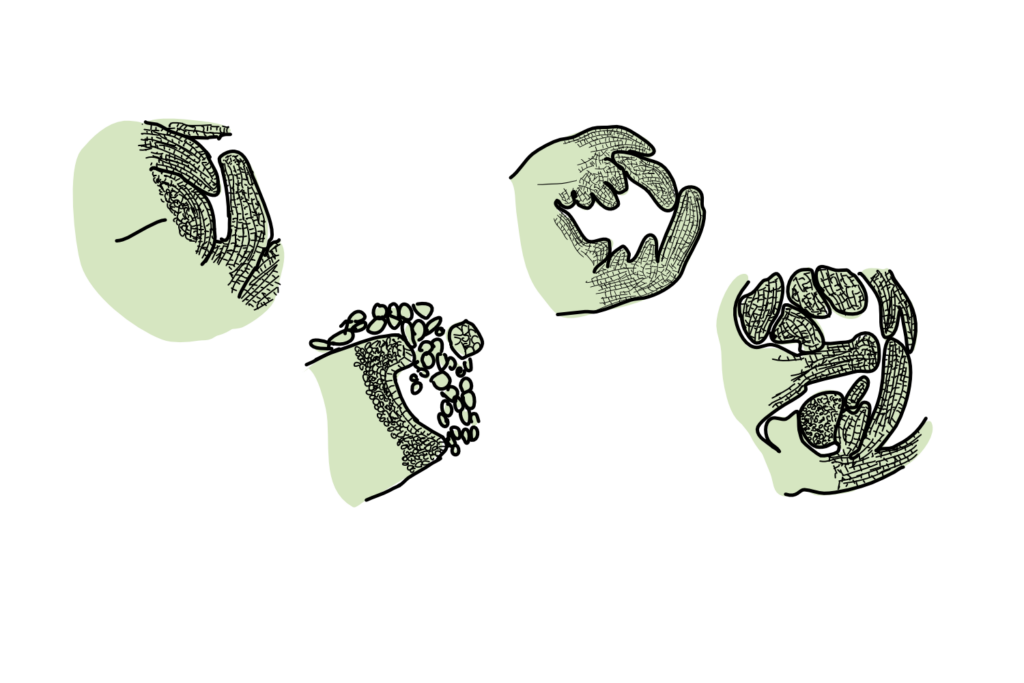
So Villar and colleagues first did some light microscopy across those many months, to define the different stages of floral development. They then undertook RNA-seq for the 4 floral development stages (S1-S4), as well as for the dormant stage (D) and compared this gene expression data to that of non-floral tissue (leaves and roots).
The scientists discovered nearly 3000 genes that were expressed at different levels depending on the stage of development. In order to narrow their search a bit, they focused specifically on transcription factors (TFs)- master regulators that run around turning other genes on and off. Working off knowledge gained in model species like Arabiodopsis, they looked particularly into the expression of MADS-box genes, which are known to be master regulators of developmental processes in plants. Eighteen different MADS-box genes were differently expressed during floral development, underlining the role of these TFs in sweet cherry floral bud development.
Along with a great deal of further analysis (check out the paper to find out more), the authors looked into the levels of certain plant hormones throughout the floral stages. This was in part because the RNA-seq showed that certain genes involved in hormone regulation changed their expression during floral development. But it was also once again working of the back of previous studies in model species like Arabiodopsis, which have shown the importance of certain hormones in regulating the floral transition.
To me, this paper provides a nice example of a seemingly contrasting idea in science: model species vs individual species. Model species are chosen because they are easy to grow, and now, after many decades of use, models like Arabidopsis have a tonne of methods and resources available for them. Generally, using a model organism, instead of your favourite exotic creature is just easier (you can read more about model species and why we use them here). Of course, at some point, it’s also necessary to move away from the models, mostly because models can’t possibly represent the diversity of life out there on the planet. But when we do journey into the unknown, we can always take the information from model species with us to help light the way.
Reference:
Villar L, Lienqueo I, Llanes A, Rojas P, Perez J, Correa F, et al. (2020) Comparative transcriptomic analysis reveals novel roles of transcription factors and hormones during the flowering induction and floral bud differentiation in sweet cherry trees (Prunus avium L. cv. Bing). PLoS ONE 15(3): e0230110. https://doi.org/10.1371/journal.pone.0230110