Evolutionary progress requires genetic material to build on- building blocks that can be shaped and changed with time and selective pressure. Plants are especially good at copy-pasting large chunks of their own DNA or even entire chromosomes, and then altering them, slowly but surely, to make new functional genes.
Sometimes however, they get fresh material from the outside.
Let’s talk about horizontal gene transfer: nature’s own way of creating GMOs.
As species move through the continuous process of evolution, their genes adapt and change. The genes can gain new functions, changes can be made to when or where the genes are expressed, or they can even be lost completely.
Genes, like the organisms they exist in, evolve through time.
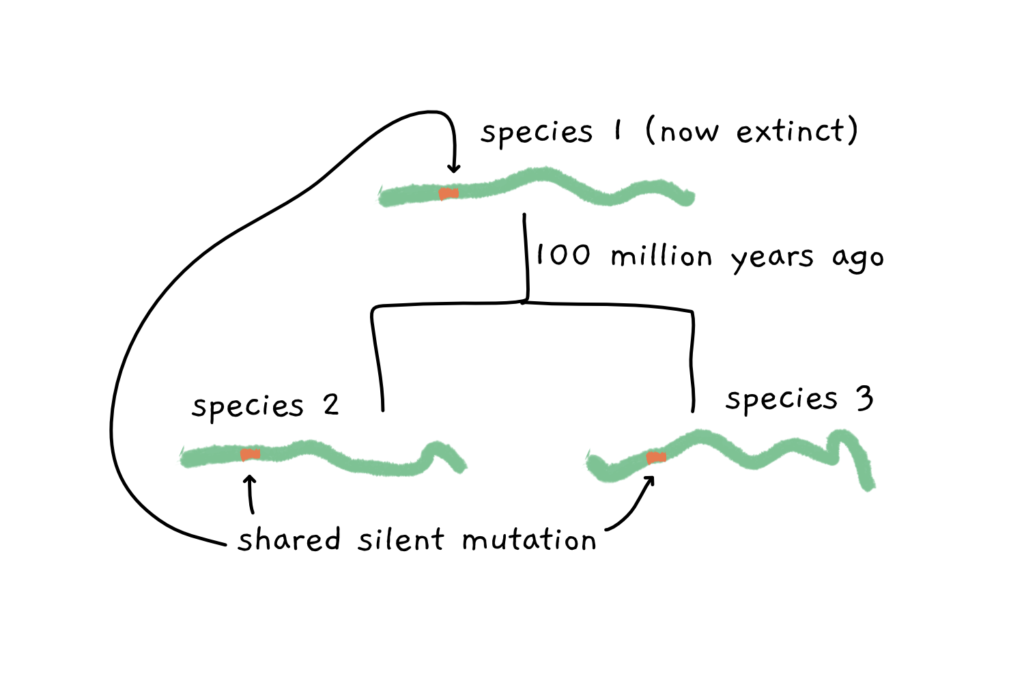
In addition to these functional changes, genes also accumulate silent mutations. These are changes to the DNA code (A, T, C and G) that don’t result in any change in the final gene product. This might be because the mutations are in parts of the DNA that don’t actually build up the protein itself. Or it might be due to the redundancy built into the amino acid genetic code: each amino acid it ‘encoded’ by a triplet of nucleotides, but they tend to have two to four triplets each. The amino acid proline, for example, is encoded by the sequence CCU, CCC, CCA or CCG, so changes in the DNA at that third position would not alter the proline-y outcome!
Anyway. Although silent mutations don’t change anything in the molecular makeup of an organism, they are very useful to track genes through evolutionary time and across diverse species species. Put simply, if a silent mutation is present in the same location in two different species, they probably share a common ancestor that acquired that mutation at one point. By doing this comparison on the entire genome with hundreds and thousands of these small differences, we can construct phylogenetic trees, mapping out the evolutionary relationship of the analysed species.
Something like this:
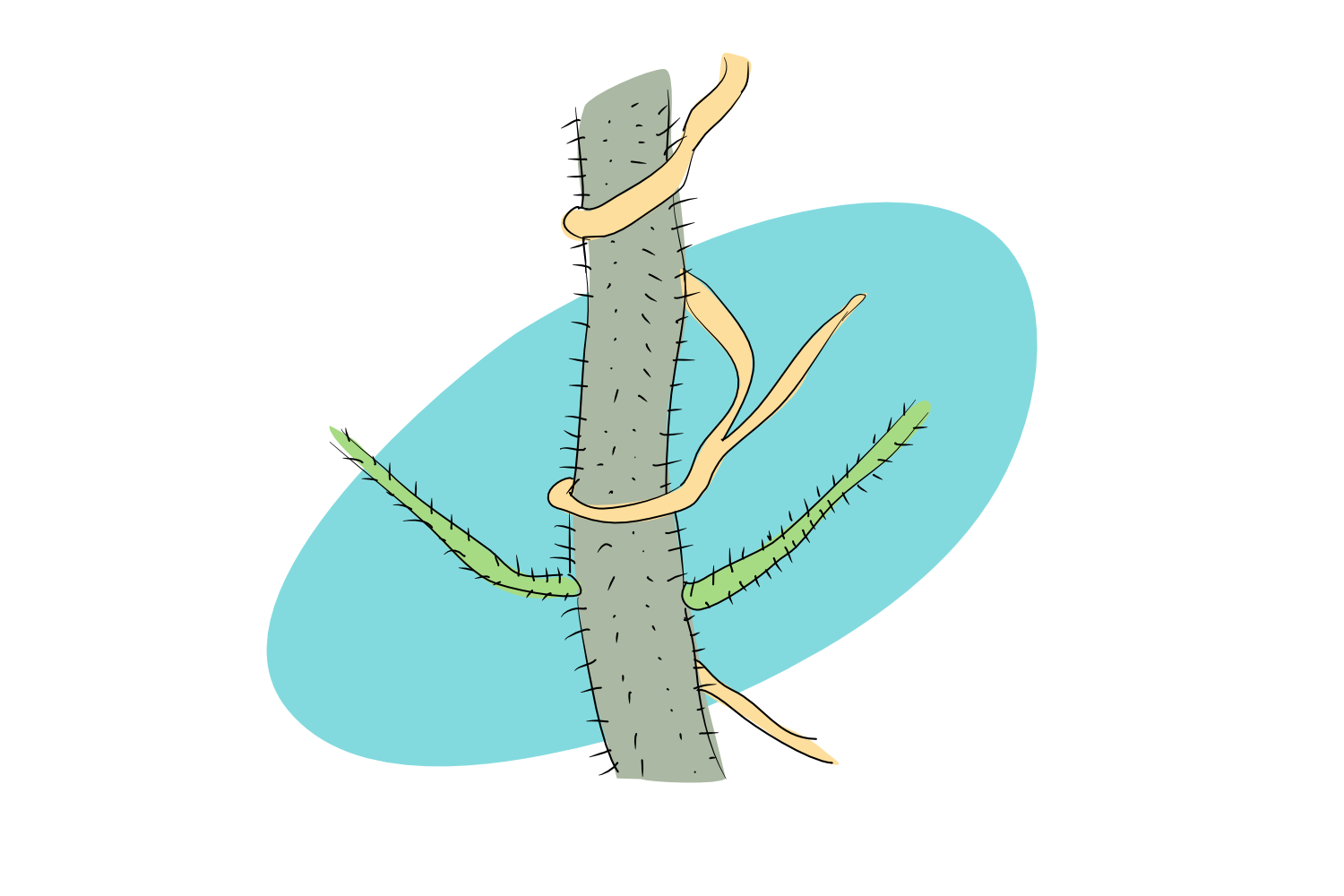
Sometimes, however, we can find pieces of the DNA puzzle that don’t fit in where we find them. Imagine a gene that is present in one plant species but in none of its close plant relatives. What’s more, there’s nothing even slightly similar to this guy in the plant genome which might suggest where the gene has evolved from.
But when you cast the net wider, you realise that there is something similar to that gene- way out there in a different set of genomes from a different set of organisms. This gene, your analysis tells you, comes from a bacteria.
So how did it get into that plant?
When two organisms, be it plant + plant, or plant + microorganism, live very close together, there’s the possibility that their genetic information will also get all close and cuddly. Messenger RNA and DNA has been shown to be able to cross cell boundaries under certain conditions. It can happen in the lab, for example, Fuentes et al. could observe entire nuclei – with all their genetic information – moving from one species to another when two species were grafted together. But it can also happen ‘in the wild’: several studies could detect the movement of mRNA and, more commonly, DNA, between parasitic plants and their hosts. To turn this exchange of genetic information into something meaningful, the receiving plant must first integrate the DNA into it’s genome, and then maintain it. At this point, we can call the occurrence a true case of horizontal gene transfer (HGT).
To this day, the role of HGT in plants is somewhat disputed. Finding evidence for HGT is not so easy: after the HGT event, the transferred and the original genes independently undergo evolutionary changes just like any other genetic information. Matching up both sequences confidently after millions of years can be quite tricky.
On top of that, genomic sequences are not always free from contamination. If some bacteria live on a plant leaf at the time of DNA extraction, chances are their DNA end up in the assembled plant genome – leading to a false clue about a non-existent HGT event. There have been several cases where more thorough analysis lead to the conclusion that hypothetical HGT events were in fact mere bacterial contamination.
Still, in a recent article, David Wickell and Fay-Wei Li present a number of examples for horizontal gene transfer in plants, an their role in evolution
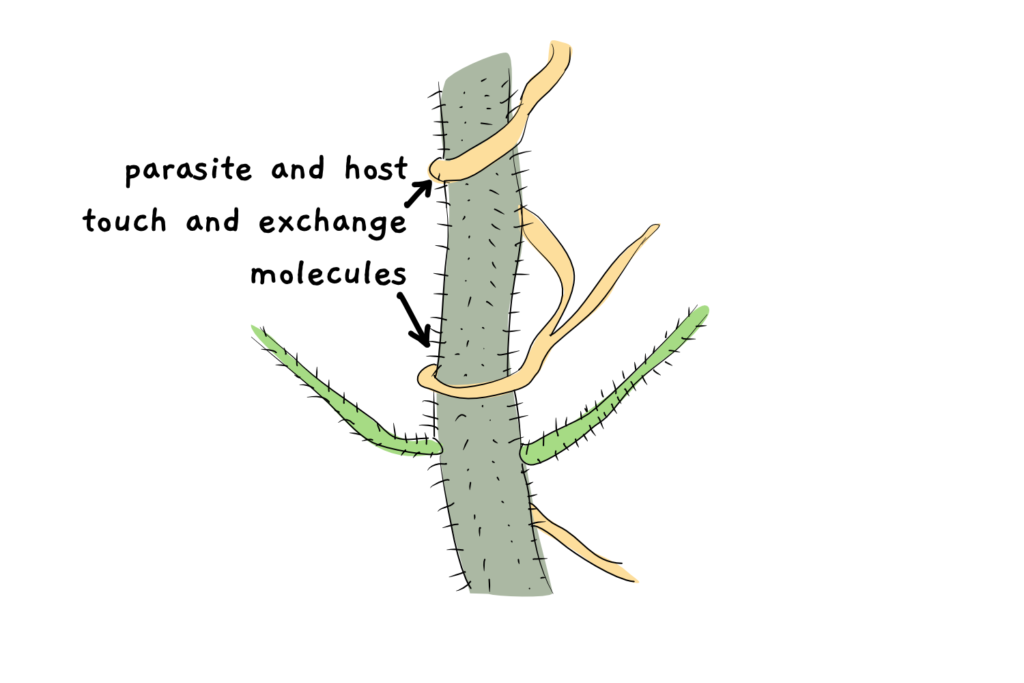
A prime example of species with HGT events are parasitic plants. They live in extremely close proximity to a host plant and therefore are very likely to get in contact with all kinds of genetic information. In the family of broomrapes (Orobanchaceae), researchers found many genes likely arising from from HGT. Some of these genes are actively expressed (i.e., they make a gene product), and several of them (18) belong to the same gene family, suggesting that there might be some functional advantage to storing these types of genes.
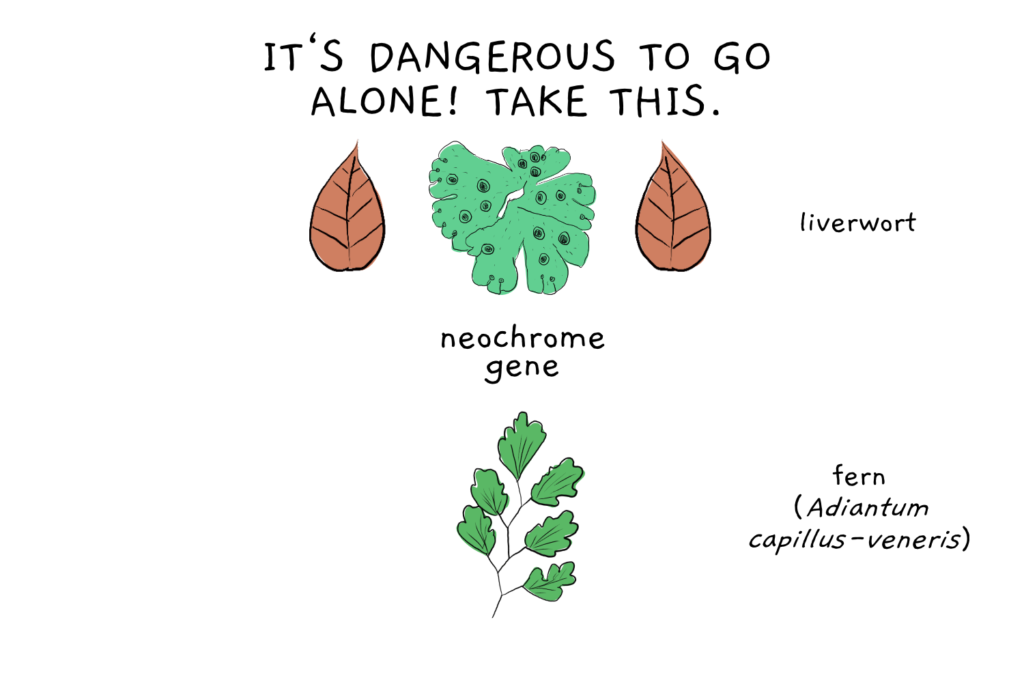
Another interesting case comes from ferns. Some fern species possess a gene encoding for neochrome, an important chimeric protein involved in adaptation to low light. When analysing related neochrome genes in liverwort, the fern version fit right in, leading to the theory that the fern neochrome gene was transfered from liverwort, probably about 178 million years ago.
Certain grasses have taken advantage of HGT as well (as much as a species can take advantage of being selected for during evolution). C4 synthesis is a way of plants to adapt their carbon fixation pathway to higher temperatures and lower humidity. The transition from “conventional” C3 synthesis to C4 occured at least 45 times independently in different species. In certain Alloteropsis species (a type of grass), key enzymes for the transition to C4 were introduced through horizontal gene transfer. The relevant genes came from other grass species, most likely through illegitimate pollination (pollen coming from the wrong species).
Why do researchers care so much about horizontal gene transfer? Well, it is a fascinating process in itself to exchange genetic information across species boundaries and to rapidly drive adaptive evolution. Essentially, HGT is nature’s way of introducing transgenes into organisms.
This also gives us a new perspective on GMOs: moving genetic information from one species to another is rare, but it isn’t new and it isn’t something that humans invented in a lab.
As with many man-made creations, nature came up with it first.
References
This article gives a very accessible overview about the state of HGT research in plants:
Wickell, D. A. and Li, F. (2019), On the evolutionary significance of horizontal gene transfers in plants. New Phytol. 🔓
This closed access publication presents the transfer of entire nuclei across graft junctions
Fuentes, I., Stegemann, S., Golczyk, H., Karcher, D., & Bock, R. (2014). Horizontal genome transfer as an asexual path to the formation of new species. Nature, 511(7508), 232–235. 🔒