You all know the mitochondria are the powerhouses of the cells.. but what exactly is required to get that power going?
Mitos were first seen as organelles way back in the 1800s, and known to be the sites of cellular respiration by the early 1900s. And although we now know that in order to do this respiration- plus a multitude of other functions- mito require thousands of types of proteins in order to function, we don’t really have a perfect idea of how many of each of those proteins is required.
That’s because, while single celled RNAseq is fast becoming a thing, single cell proteomics just isn’t quite there yet- the machines aren’t powerful enough. So in order to answer complex questions like ‘how many protein are in a mito’, as well as the slightly more philosophical ‘how many mito does it take to power a lightbulb?’, Philippe Fuchs, Nils Rugen and their colleagues had to go at it the hard way.
Before you read more, see if you can guess how many proteins are in a mitochondrion!
The procedure looked a bit like this. The scientists isolated mitochondria from plant cells, and then further isolated all of the proteins from those mitochondria. The proteins were digested – chewed up into smaller pieces called peptides – that could then be measured by a common method known as mass spectrometer.
The researchers took great care in assuring that this data was of the highest quality, so that they could interpret it in a quantitative way. A whole lot of extra calculations later, involving integration of known data from the scientific literature, and a few assumptions, and the authors could present an amazing work: they could approximate how many of each protein are required to make a single functional mitochondrion.
There are certain assumptions that had to be made, as well as certain intrinsic limitations associated with the technology used, so let’s get those out of the way before we get to the key findings of the work.
Assumption 1: That a cultured mitochondrion is an average mitochondrion
The authors purified mitochondria from dark-grown Arabidopsis cell cultures, as opposed to those taken from whole plants. The suspensions are convenient, and most importantly, growth in darkness means that there aren’t so many chloroplast proteins (normally present in huge amounts) that could contaminate the work. Of course, the authors are quick to point out that certain functions might be underrepresented in their system. For example, they noticed that the accumulations of proteins involved in photorespiration – a process required to clean up chloroplast mistakes – was pretty low compared to those expected in a light-grown plant.
Assumption 2: That their mitochondria were pure
Look, purified mitochondria are never completely pure. When you pull, or more accurately, spin, the mito out of their cellular context, they always bring a few extra, sticky proteins with them. Across their four replicate experiments, the authors identified nearly 3000 unique proteins in their isolated mitochondria.
Although only about 1/3 of the identified proteins were true mitochondrial proteins (917 proteins), those proteins represented the bulk of the protein molecules – 84%. Meaning that those extra 2000 or so individuals were mostly present as just a couple of lonely molecules, hanging onto the sides of the mito. We should note that these kinds of experiments generally use a cutoff – in which proteins with lower numbers are simply removed as ‘noise’. But the authors didn’t want to risk losing true mitochondrial proteins that might be present at low abundances, so they kept their cutting-off to a minimum!
Assumption 3: That the mass spec and iBAQ are truly quantitative
The authors used mass spec, followed by a method known as iBAQ to first identify and then calculate the number, of each kind of protein. iBAQ stands for ‘intensity-based absolute quantification’, and it’s a way of calculating protein amount based on the intensity of that protein’s peptides. Unfortunately, as with all methods, iBAQ isn’t quite perfect. Different peptides have different biochemical properties, and these properties can change how well they are fed into the machine and detected. Proteins with small size and high hydrophobicity, for example, are generally underrepresented. And the authors saw evidence of this in their study. Two small hydrophobic proteins that should be in the mito (Atp9 and MatR), could not be detected, while other hydrophobic proteins were found in lower amounts compared to non-hydrophobic proteins belonging to the same complex (and known to be present in equal amounts). So some proteins are almost definitely underrepresented, but for now, iBAQ is the best option we have.

Assumption 4: That mitochondria are kind of round
In order to get an approximate number of proteins per mitochondrion, the authors first had to define what exactly a mitochondrion is. Which is not as easy as it sounds. As seen in microscope images, flowering plants like Arabidopsis contain mitochondria which are, on average, roughly bean shaped and between 0.4 and 2.0 µm long. But on any normal day, those tiny beans are undergoing constant fusion and fission, exchanging goods and services, and changing shape and size. It’s given them the nickname ‘discontinuous whole’ (which I never not think of as ‘discontinuous hole, which is a whole new riddle). Anyway, in order to make their calculations, the authors had to go for an average – a basically round sphere with 0.8 µm diameter.
Assumption 5: That mitochondria are 25% protein, by weight… and a bit more math
Knowing the size and (from other people’s experiments) the density of the mitochondria allows calculation of each mito’s approximate weight. Which comes in at 322 femtograms. Femto- means x 10-14, so that’s just 0.000000000000322 g. If the mitochondrion is 25% protein by weight (a number also taken from the literature), then that makes 80.4 fg of protein per mitochondrion.
Knowing the approximate weight of the average mitochondrial protein means that those femtograms could be used to calculate an estimated total number of proteins in a single mitochondrion. The number of each individual protein was then calculated by multiplying that total protein number, by the proportional value worked out by iBAQ. Et voilà. You’re done!
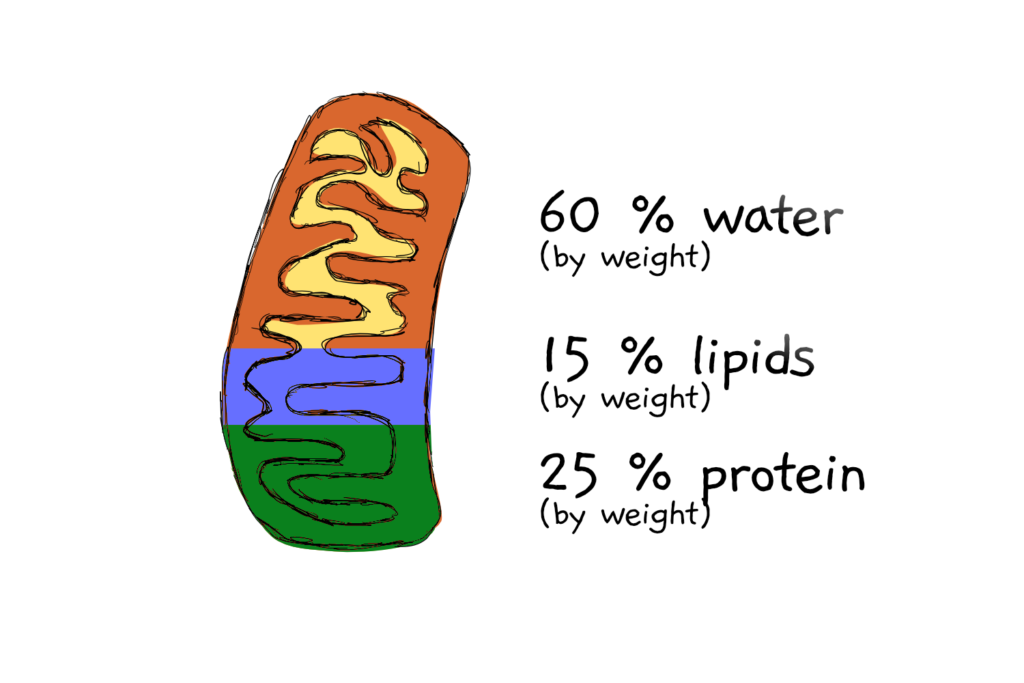
As you can probably tell, getting to the final numbers was no mean feat. As joint-first author Philippe Fuchs stated:
You will have noticed that our work originated from a rather big collaborative effort involving a number of independent research groups. To reach our goal, i.e. calculating the copy numbers of individual proteins per single mitochondrion, we are relying on a number of theoretical and experimental considerations. One of the major challenges was to collect, share and discuss these considerations with all the authors so that I could apply them as appropriately as possible before we started to actually crunch through the numbers. Since our work was more a community effort rather than a project controlled and guided by one research group, synchronising these considerations was much more complex and challenging than what I anticipated.
So now that the scientists have done the hard part, I want to share some of the cooler findings from the paper. Let’s begin shall we?
There are 1.4 million protein molecules in a mitochondrion!
I’m just going to let that number sit there. One point Four MILLION.
By the way this number is remarkably similar to a previous estimate, using completely different assumptions and calculations, of 1.6 million proteins per mito. We want to note that, for the most part, the authors found similar numbers to those previously suggested (again, all by different methods). Which is a reassurance that their assumptions and calculations were pretty reasonable.
The protein with the highest copy number… is VDAC1!
If I had to guess, I would assume it was something involved in oxidative phosphorylation (OXPHOS!). You know, those membrane complexes that make most of the ATP (energy) that the cell uses to literally do everything?!
But no, VDAC1 is the grand winner, with 44 thousand copies found. VDAC1, stands for Voltage-dependent anion channel 1, and as the name suggests, it’s involved in moving small metabolites – required for all kinds of biochemical pathways- across the outer mitochondrial membrane. Four other VDAC isoforms (VDAC 2-5) were also pretty abundant, and together the VDACs made up more than 80k protein molecules. Based on the approximate size of the VDACs themselves, and the calculated surface area of the outer membrane, this means that VDACs take up more than 1/3rd of the total surface area of the membrane.
Our findings gave me an entirely new perspective … Consider that VDACs are controllable channels, I think the outer mitochondrial membrane is much more an adjustable nano-sieve rather than a simple lipid bilayer. This in turn could suggest that the outer mitochondrial membrane is directly involved in regulating the metabolic activity of a mitochondrion by simply adjusting the provision of metabolites.” P.F, author
Interestingly, transporters also made up a pretty big chunk of the proteins in the inner mitochondrial membrane, occupying about 10% of the total membrane area. Again suggesting the importance of import (and export) in controlling mito metabolism!
But… it’s still all about ATP
In total, a bit under half of all the mitochondrial proteins are somehow involved in the production of ATP. The four complexes (I-IV) involved in oxidative phosphorylation, the final pathway of cellular respiration make up 220 thousand proteins alone, and probably occupy about 10% of the entire invaginated inner mitochondrial membrane.
ATP synthase takes up another 8% of that space. The protein complexes are not found in equal amounts – Complex I, II, IV, and the electron carrier Cytochrome c had about 2-3 thousand copies each, while Complex III and the ATP synthase had nearly 6.5 thousand copies.
Over in the mitochondrial matrix, there’s a similar story. The proteins involved in performing the 10 enzymatic steps of the TCA cycle represent about 80% of the protein volume of the matrix!
We now know how many mito it takes to change, I mean power, a lightbulb
In many instances, our findings were not surprising and confirmed findings from previous studies. However, calculating the abundance in the form copy numbers rather than for instance classifying a protein as ‘moderately abundant’ provided us the resources to apply another step of calculations, e.g. […] the catalytic capacity of a certain process in a single mitochondrion. These calculations may support and shape our understanding of certain processes/proteins in mitochondria. P.F, author
One of the coolest of these calculations is that of the approximate volume of mitochondria required to power a light bulb. Assuming that 200 nanomol of oxygen can be processed per minute per gram of mitochondrial protein, and a ADP to Oxygen ratio of 2.5, then a single mito makes nearly 1 million ATP molecules per second (or 151 ATPs/ATP-synthase/second). This means that less than 20 ml (a shot from a stingy bar keeper) would be enough to power a standard household 7W LED bulb!

The world almost definitely has more TOMs than TIMs
Nearly 12% of protein molecules found were involved in protein import, as well as processing (maturation, folding and degradation) – more than 200k protein molecules in total. Maybe not surprising, given that most of the mitochondrial proteome has to be made in the cytosol form nuclear-encoded genes, and then brought into the mito.
Interestingly, the protein channels for the outer membrane, called TOMs (Translocase of the Outer Membrane) were much more abundant than those for the inner membrane (TIMs). Assuming that the number of TIMs is the limiting factor in protein import, and given a calculated average protein length of 353 amino acids and assumed translocation rate of 40 amino acids/second, means that the number of proteins in the mito could be doubled in just six and a half hours!

Facts, figures… function!
One interesting side-effect of the precise calculations means that abundance-based hypothesizes could be made about protein function. For example, in plants, heme biosynthesis largely occurs in the plastid, but there has been some debate over whether the last step of the pathway – the chelation of iron – might go down in the mitos. Relatively high copy numbers of the enzyme responsible for this last step found by the authors supports this case.
In another example, the authors examined the nearly 300 members of the mitochondrial-targeted PPR protein group – a family mostly involved in RNA processing. They could find only 100 or so of these PPRs, likely because the remaining accumulated to such low levels that they couldn’t be detected, and the majority of those found had less than 20 copies. With several exceptions. Many PPRs are involved in highly specific interactions – one PPR to one mRNA, for example. But some PPRs are involved either in more general protein–protein interactions, or match up with the much more abundant ribosomal RNA molecules. Unsurprisingly, these PPRs were found at higher levels than their cousins. Similarly, a protein known as DYW2 has long been suggested to be involved in a PPR-requiring process known as RNA editing- not as a specific factor, but as a more general editing enzyme. The relatively high number of these molecules compared with other editing PPRs seems to support this role.
Substochiometry, fission, fusions
Many PPRs, as well as other proteins, were found in substochiometric amounts, meaning there was, on average, less than one copy per mitochondrion. As joint-first author Nils Rugen stated:
I was surprised to see that many proteins are of substoichiometric abundance, meaning that, at a given time point, some mitochondria indeed carry these proteins whereas others do not. The constant fusion and fission processes taking place within the cellular population of mitochondria then ensure the equal distribution of these proteins over time.
The idea that mitochondria must constantly undergo fusion and fission in order to function was further suggested by the low number of DNA binding proteins, which indicates the average presence of the mitochondrial genome in just one of every three mitochondria!
So those are our favourite facts from an amazing paper. When we asked the authors what comes next, they pointed to the arrival of increasingly better technology, as a method to both increase the chances of picking up rare proteins, and to better understand mitochondrial heterogeneity:
We surely overlooked quite a few mitochondrial proteins due to the detection limits of our currents mass spectrometers. These machines are currently evolving fast to become more and more sensitive. N.R
I am really looking forward to see the next generation of mass spectrometry being applied to determine the protein inventory of single organelles. Our work focussed on an average mitochondrion since there are currently no practical means to assess the heterogeneity of organelle populations. P.F.
Even though we think of cells as mostly water, studies like this one remind us time and time again how densely packed these tiny biological compartments are. Literally millions of individual protein units sit side by side, performing complicated biochemical reactions, that ultimately make up the incredible lifeforms of this planet.
Next up- has someone (some many ones) got a few years time to do this for the chloroplast?
References:
This article is based on:
Fuchs P, Rugen N, Carrie C, Elsasser M, Finkemeier I, Giese J, Hildebrandt TM, Kuhn K, Maurino VG, Ruberti C, Schallenberg-Rudinger M, Steinbeck J, Braun HP, Eubel H, Meyer EH, Muller-Schussele SJ, Schwarzlander M. 2019. Single organelle function and organization as estimated from Arabidopsis mitochondrial proteomics. Plant Journal.
Acknowledgements: A huge thank you to Philippe and Nils for answering our questions and providing us with quotes! Double thanks for doing it in holiday time!
Disclaimer: We have met, know, and even like some of the authors involved in this paper. I actually chose this paper based on the title and a quick read of the abstract before looking at the names, but we thought you should know. We also have a favourite. It is E.M.